Pythong-based neuroimaging visualization in R
Publish date: April 10, 1019
```{r setup}
knitr::opts_chunk$set(echo = FALSE)
knitr::knit_engines$set(python=reticulate::eng_python)
library(knitr)
library(reticulate)
use_python("/anaconda3/bin/python3.6")
```Running nilearn/pysurfer visulization using pythong code
Prerequisite: Install python, nilearn (https://nilearn.github.io/) and pysurfer (https://pysurfer.github.io/index.html)
The version of python has to be correctly specified using use_python().
## /anaconda3/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.
## from ._conv import register_converters as _register_converters## /Users/seonjoolee/nilearn_data/neurovault/collection_658/image_10426.nii.gz
```{python,message=FALSE,qietly=T}
from nilearn import datasets
motor_images = datasets.fetch_neurovault_motor_task()
stat_img = motor_images.images[0]
print(stat_img)
from nilearn import plotting
plotting.plot_glass_brain(stat_img, threshold=3,output_file='test1.png')
```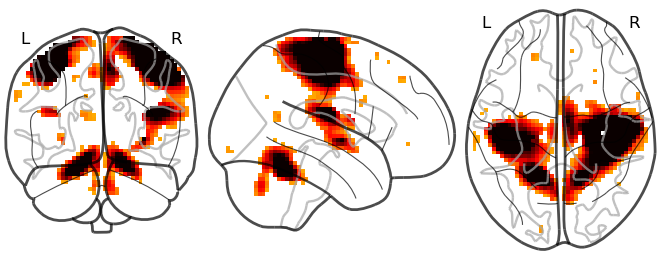
Glass brain visualization
pysurfer visualization
1. First option: Calling python objects in R
- nib\(freesurfer\)read_annot function does not work properly.
```{r,echo=F, quietly=F}
os=import("os")
mlab=import("mayavi")
nib=import("nibabel")
brain=import("surfer")
subject_id = 'fsaverage'
hemi = 'lh'
surf = 'inflated'
## The following line caused trouble...
# nib$freesurfer$read_annot('/Applications/freesurfer/subjects/fsaverage/label/lh.aparc.annot')
a<-brain$Brain(subject_id, hemi, surf,subjects_dir='/Applications/freesurfer/subjects')
a$add_text(0.5,0.5,'Test','test')
a$save_image('Test.png')
a$close()
rm(list=ls())
```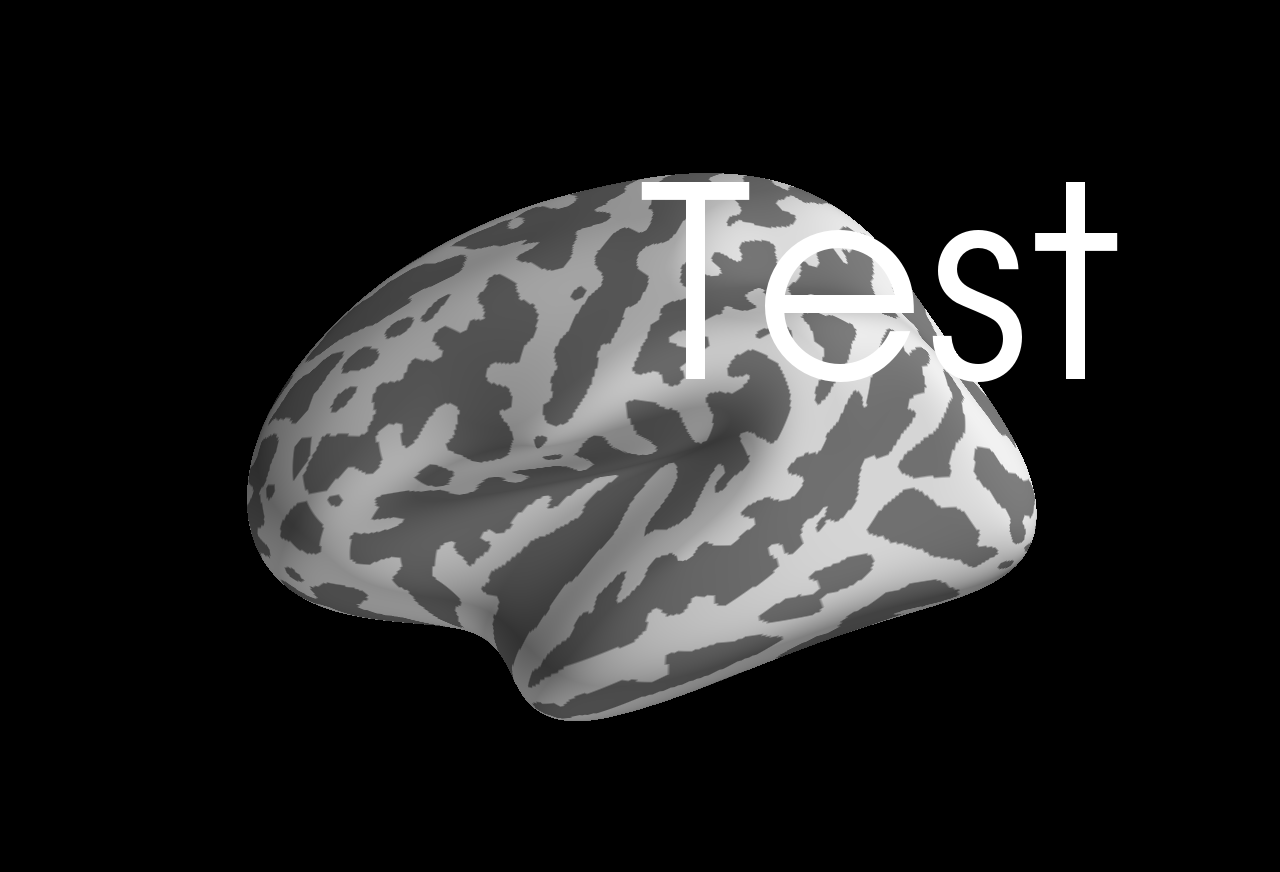
Example file
2. Run python code directly
Runs smoothly.
To pass data between R and python, we should use py object
```{r}
py$subject_id = "fsaverage"
py$hemi = "lh"
py$surf = "inflated"
```## colormap sequential: [5.00e-01, 6.25e-01, 7.50e-01] (opaque)```{python}
import os
import numpy as np
import nibabel as nib
from surfer import Brain
br = Brain("fsaverage", "lh", "inflated",subjects_dir='/Applications/freesurfer/subjects')
br.add_text(0.5,0.5,'Test','test')
aparc_file = os.path.join('/Applications/freesurfer/subjects',
subject_id, "label",
hemi + ".aparc.a2009s.annot")
labels, ctab, names = nib.freesurfer.read_annot(aparc_file)
rs = np.random.RandomState(4)
roi_data = rs.uniform(.5, .8, size=len(names))
vtx_data = roi_data[labels]
vtx_data[labels == -1] = -1
br.add_data(vtx_data, .5, .75, thresh=0, colormap="rocket", alpha=.8)
br.save_image('test2.png')
br.close()
```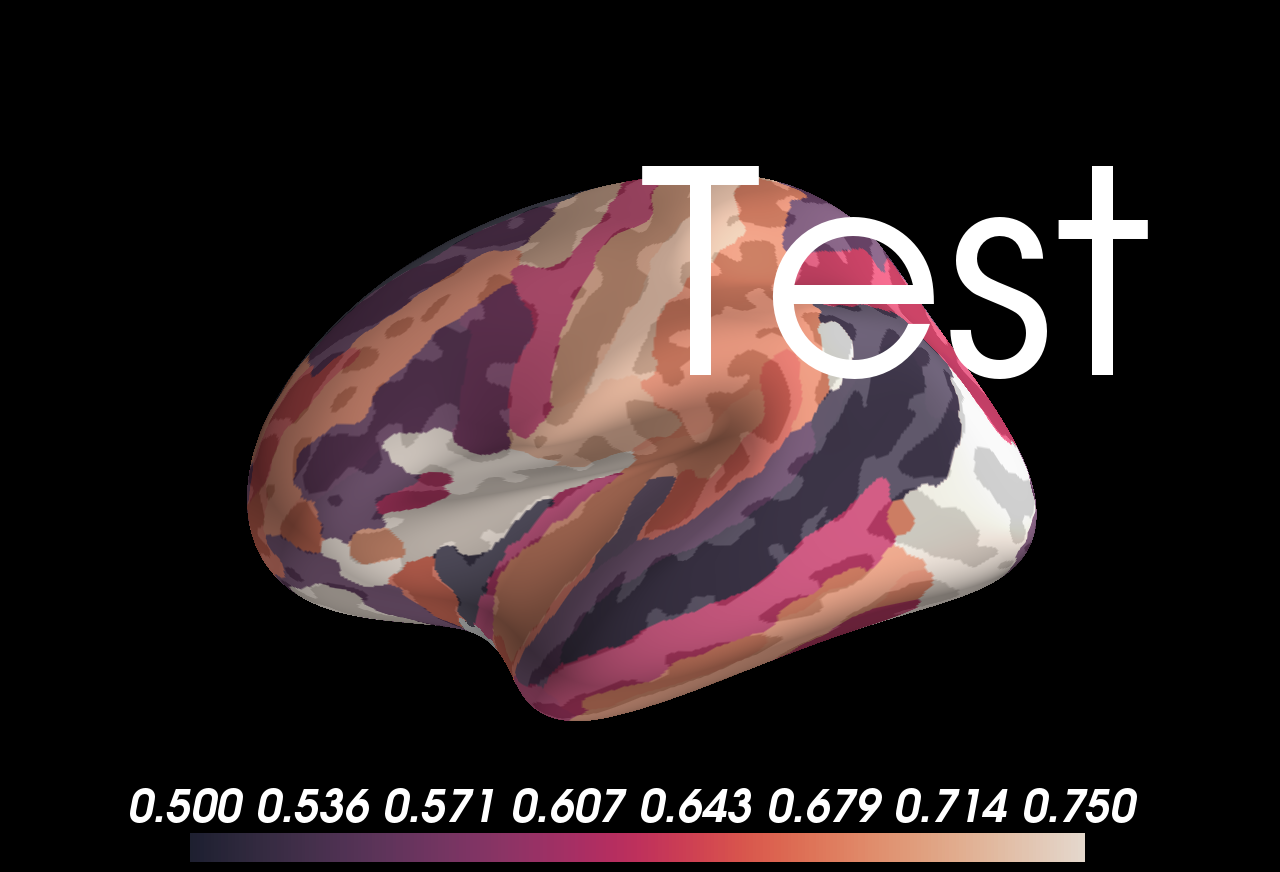
ROI-level values overlaid on the aparc 2009
Rmd file
Available in the following link: https://drive.google.com/open?id=1p70SfXJZ7BeQBM2dxbBMsAqaQgwP5hsQ
This version is very preliminary. Any suggestion is welcome!
