Introduction to Independent Component Analysis
Introduction to Independent Component Analysis with fMRI examples
Denition of ICA
\[X = AS\]
\(X\) is a random \(M \times T\) matrix representing the observed matrix at T time points
\(S\) is a random \(M \times T\) matrix where each row is mutually independent
A is an M \(\times\) M full-rank non-random mixing matrix
Estimate the unmixing matrix \(W = A^{-1}\) such that \(\hat{S}=\hat{W}X\) is the estimates of \(S\).
ICA Assumption 1: Independence
Definition and fundamental properties: we define that two variables and are independent if and only if the joint pdf is factorizable in the following way: \[p(s_1,s_2 )=p(s_1) p(s_2)\]
Expected Value Definition (came form moment generating function):
\[E(h_1(s_1)h_2(s_2))=E(h_1(s_1)) E(h_2(s_2))\] for any functions \(h_1\) and \(h_2\) such that the above expected values exist and are well-defined.
2: Non-Gaussian
- Why Gaussian variables are forbidden? the distribution of any orthogonal transformation of the Gaussian \((s_1,s_2)\) has exactly the same distribution as \((s_1,s_2)\).
- If \(R\) were some arbitrary orthogonal matrix so that \(RR^{\top}=R^{\top}R=I\). Let \(A^{\prime} = AR\). If data had been mixed according to \(A^{\prime}\) (instead of \(A\)), we would have \(X\) as a Gaussian with a mean still 0 and covariance: \[ Cov(X)=E XX^{\top}=E A^{\prime}SS^{\top}{A^{\prime}}^{\top}\]
\[ E ARSS^{\top}R^{\top}A^{\top}=ARR^{\top}A^{\top}=AA^{\top}\]
Measure of Non-gaussianity
We need to have a quantitative measure of non-gaussianity for ICA Estimation.
Kurtosis : gauss=0 (sensitive to outliers) \[kurt(Y)=E Y^4 - 3 (EY^2)^2\]
Entropy : gauss=largest \[H_Y(y)=-\int f_Y(y)\log f_Y(y)dy\]
Neg-entropy : gauss = 0 \[J(Y)=H(Y_{gaussian})-H(Y)\]
Approximations \[J(Y) \approx \frac{1}{12}(EY^2)^2 +\frac{1}{48} kurt(Y)^2\]
Caution & Limitation
Scaling
For any constant \(a\), \[ X=AS=aA \cdot \frac{1}{a} S\]
It is often assumed that each source has zero mean and unit variance.
2. Signal Permutations
- The order of mixing matrix and independent components are unknown.
\[ X=AS=AP^{-1} PS\]
3: Number of Sensors
- The number of separated signals cannot be larger than the number of inputs.
- Current research is being done to reduce this constraint.
Example: Data generation
- Two soures were generated from standard normal and Uniform(0,2\(\sqrt{3}\)).
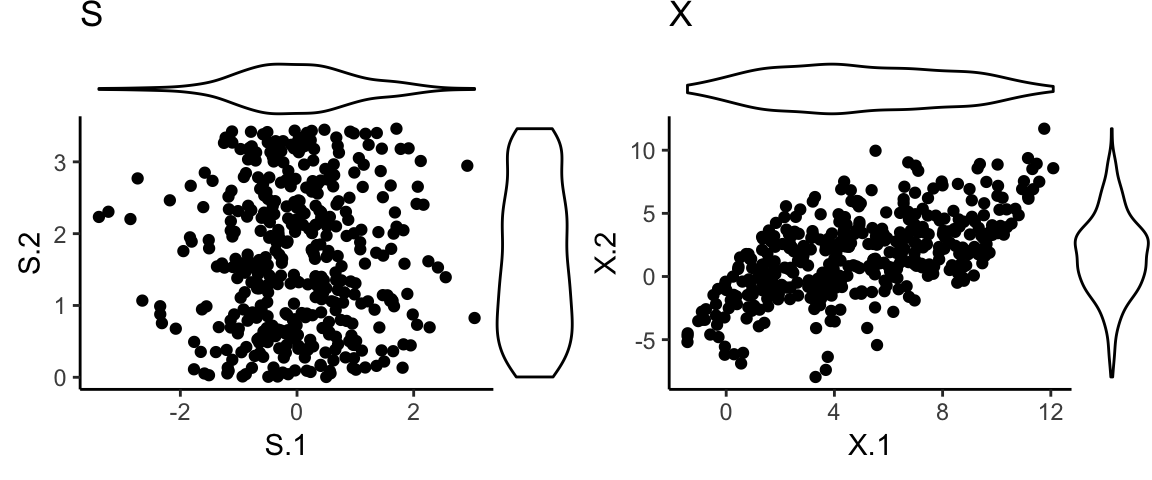
Example: Source separation
- PCA vs. ICA
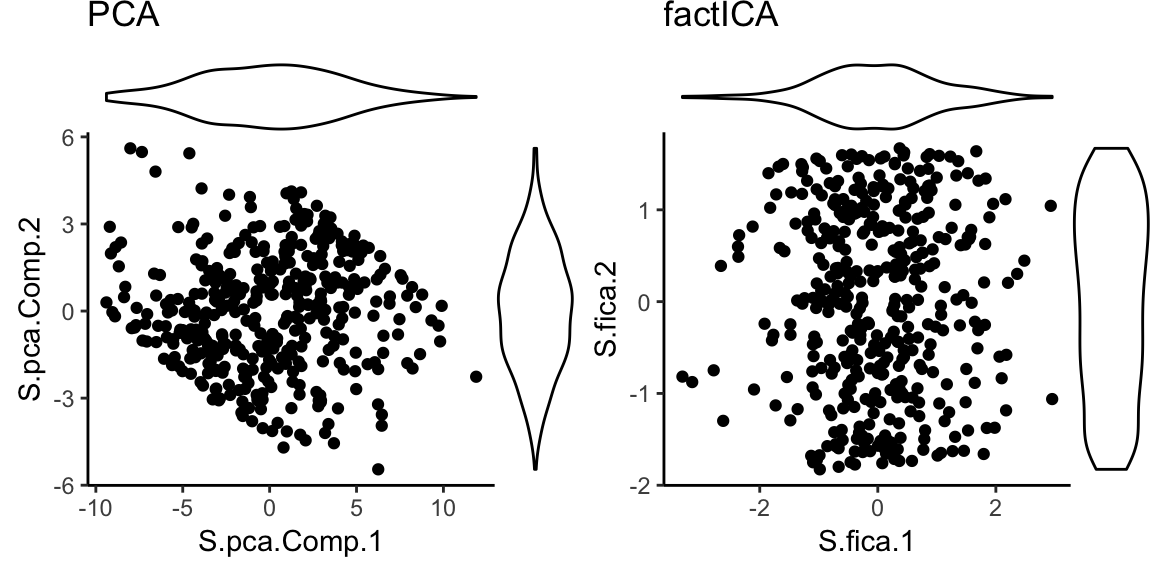
Example: Source separation (Cont.)
Why PCA could not recover sources while ICA could?
PCA finds the directions that explains most variance (2nd order), while ICA finds the directions that maximize “independence”.
If the soures are from normal distribution, finding direction that makes the estimated \(S\) uncorrelated will achieve finding independent sources.
However, if the sources are not from gaussian distribution, uncorrelation does not mean independence.
e.g. S1 ~ unif(-1,1)*sqrt(6), S2 ~ exp(1)
X1=2*S1 + S2; X2=S1-S2: X1 and X2 are uncorrelated, but not independent.
(X1-X2, X1+2X2) = (S1+2S2, 4S1-S2), Uncorrelated, but not independent.
functional Magnetic Resonance Imaging (fMRI)
Creates a series of images that capture blood oxygination level dependent (BOLD) signal in parts of the brain
Non-invasive method
Popularly used in clinical practice and human brain research
-Find the part of the brain handling certain functions such as movement, sensation, thought, etc.
- Characterize the brain functions
/presentation/p2011/defense/pictures/homer_braincolor2.jpg)
/presentation/p2011/defense/pictures/homer_mri.jpg)
functional Magnetic Resonance Imaging (fMRI) Data
/presentation/p2011/defense/pictures/three.png)
- fMRI volume image. Red and yellow area highlight activated
fMRI Data Matrix
/presentation/p2011/defense/pictures/intro_imagetomat2.png)
fMRI Data Decomposition
\[X = AS \]
/presentation/p2011/defense/pictures/intro_mixture2.png)
/presentation/p2011/defense/pictures/intro_spmap3.png)
ICA in fMRI
Relying on the intrinsic structure of the data
No assumptions about the form of the HRF or the possible causes of responses are made.
The only assumption is mutual independence in “Space” (spatial ICA) or in “Time” (temporal ICA).
Temporal vs. Spatial ICA}
/presentation/p2016/snu2016/tsICA.png)
Temporal vs. Spatial ICA
Temporal ICA
Components have independent temporal dynamics: “Strength of one component at a particular moment in time does not provide information on the strength of other components at that moment”
Components may be correlated/dependent in space
Popular for cocktail party problem or EEG
Spatial ICA
- Components have independent spatial distributions: “Strength of one component in particular voxel does not provide information on the strength of other components in that voxel”
- Components may be correlated/dependent in time
- Popular for fMRI
Real example data
Dataset was downloaded from FSL (https://fsl.fmrib.ox.ac.uk/fslcourse/)
Demostrate run ICA in fsl and R, and how to look at the results.
Dataset was “preprocessed” using FSL FEAT (default setting)
library(fslr)
img = readnii('ica_intro_fsldata/filtered_func_data.nii.gz');
print(img)## NIfTI-1 format
## Type : nifti
## Data Type : 4 (INT16)
## Bits per Pixel : 16
## Slice Code : 0 (Unknown)
## Intent Code : 0 (None)
## Qform Code : 1 (Scanner_Anat)
## Sform Code : 1 (Scanner_Anat)
## Dimension : 64 x 64 x 21 x 100
## Pixel Dimension : 4 x 4 x 6 x 1
## Voxel Units : mm
## Time Units : secOne volume

Five random voxels
ts.plot(t(img[c(25,30,35,40,45),32,10,]),col=1:5)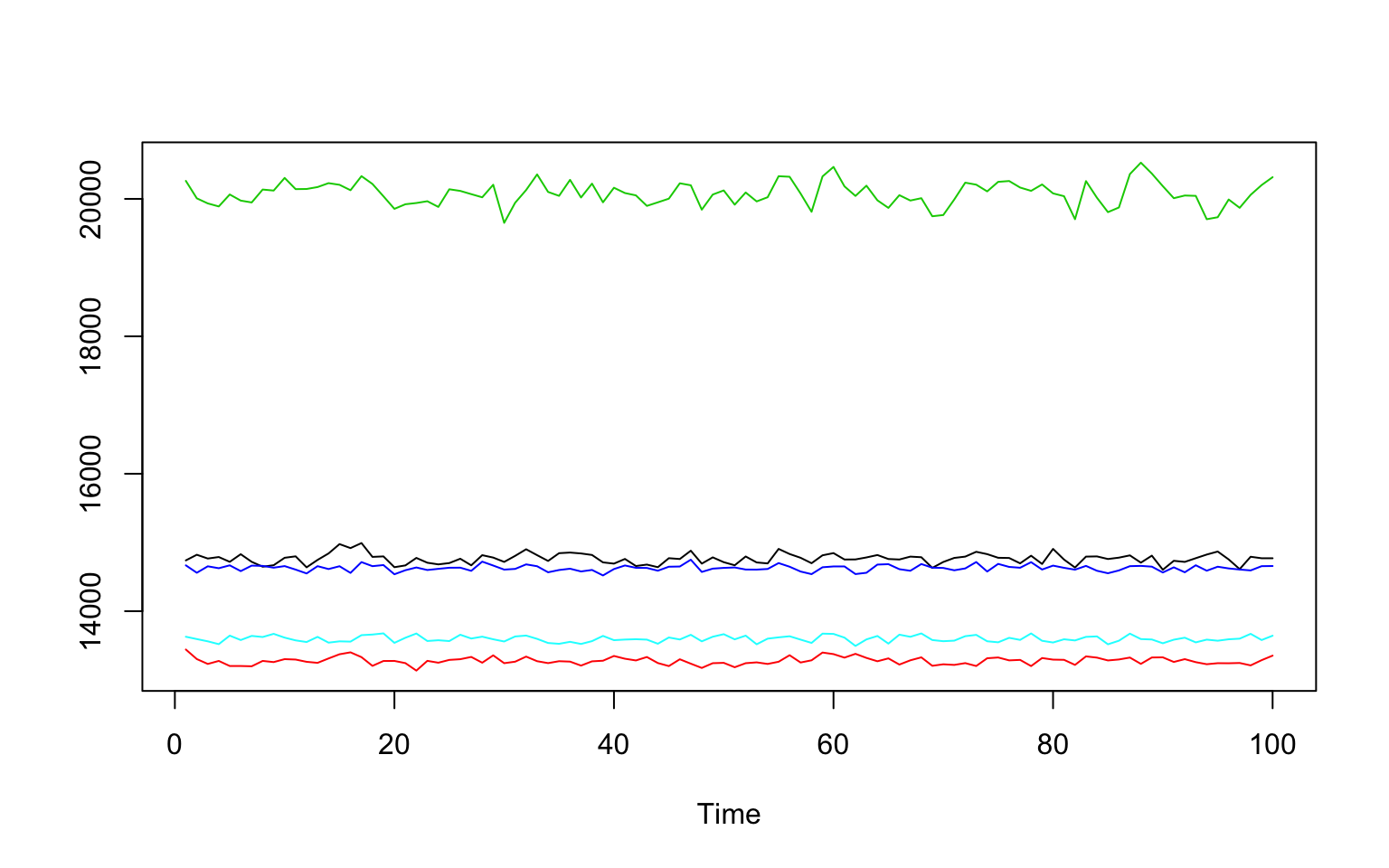
Five random voxels
ts.plot(scale(t(img[c(25,30,35,40,45),32,10,])),col=1:5)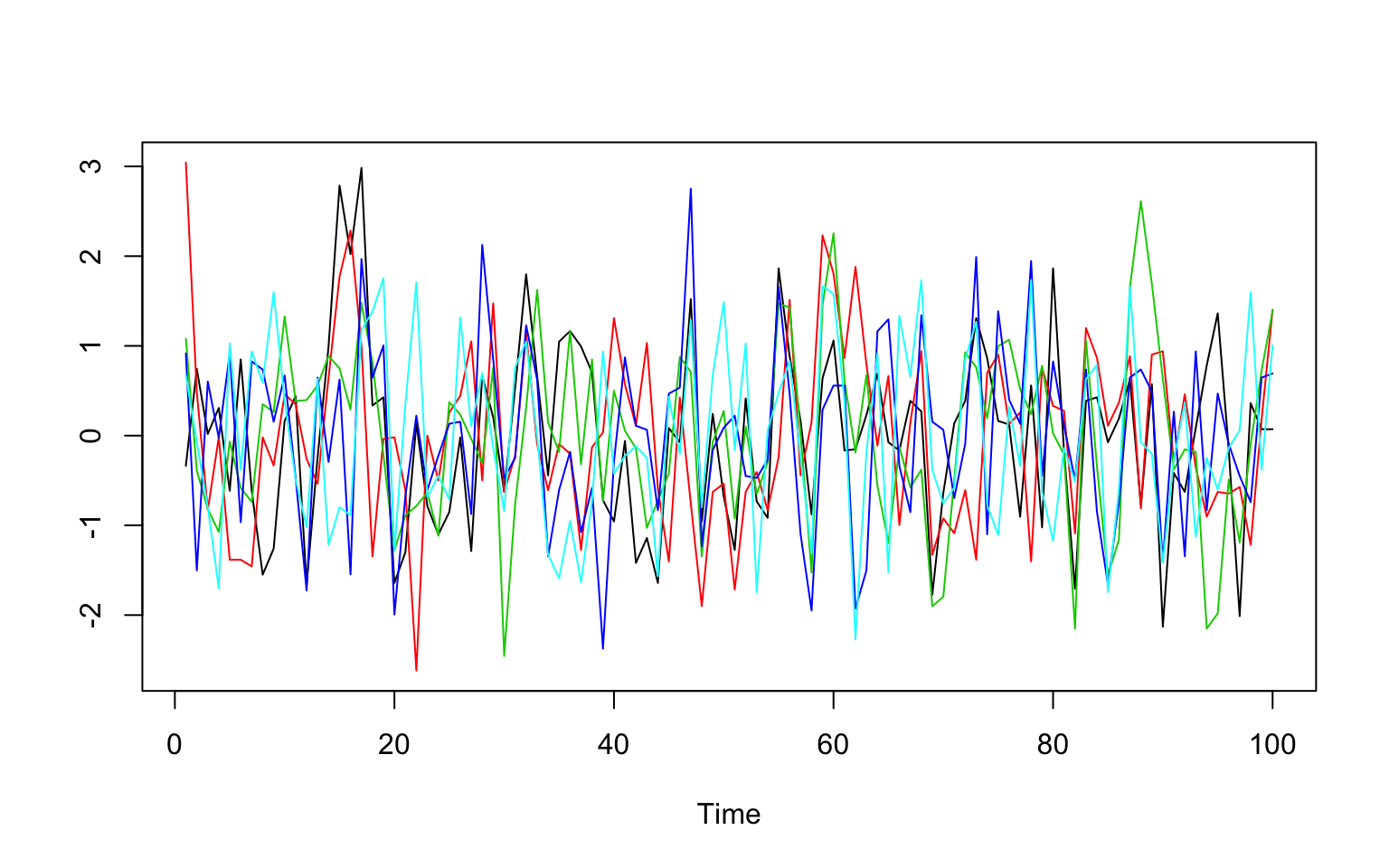
How to apply ICA
The easiest way is to use a neuroimage package such as FSL-Melodic, GIFT, etc.
In R, f.ica.fmri function is available in AnalyzeFMRI R package.
Typically spatial ICA is applied for FMRI, and temporal ICA is applied to EEG.
The basic steps include: array flattening, prewhitening, optimization (estiomation), spatial/temporal component recovery
Practice (Spatial ICA):
- Flattening 4D volumes to 2D (V \(\times\) T) matrix
dims=dim(img)
X=t(array(img, dim=c(dims[1]*dims[2]*dims[3],dims[4])))
X.c = X - apply(X,1,mean)Prewhitening
Select number of components (in fMRI application it is often set to be 20, but there’s also some systematic way to determine).
Sigma.eigen=svd(X.c)
n.comp=10
Y = t(Sigma.eigen$v[,1:n.comp]) spatial ICA example (Cont.)
- ICA is applied to Y
set.seed(1234)
system.time(re<-fastICA(t(Y),10))## user system elapsed
## 2.592 0.871 3.472temporal component recoveray
A.est = Sigma.eigen$u[,1:n.comp] %*% re$A
ts.plot(scale(A.est),col=1:10)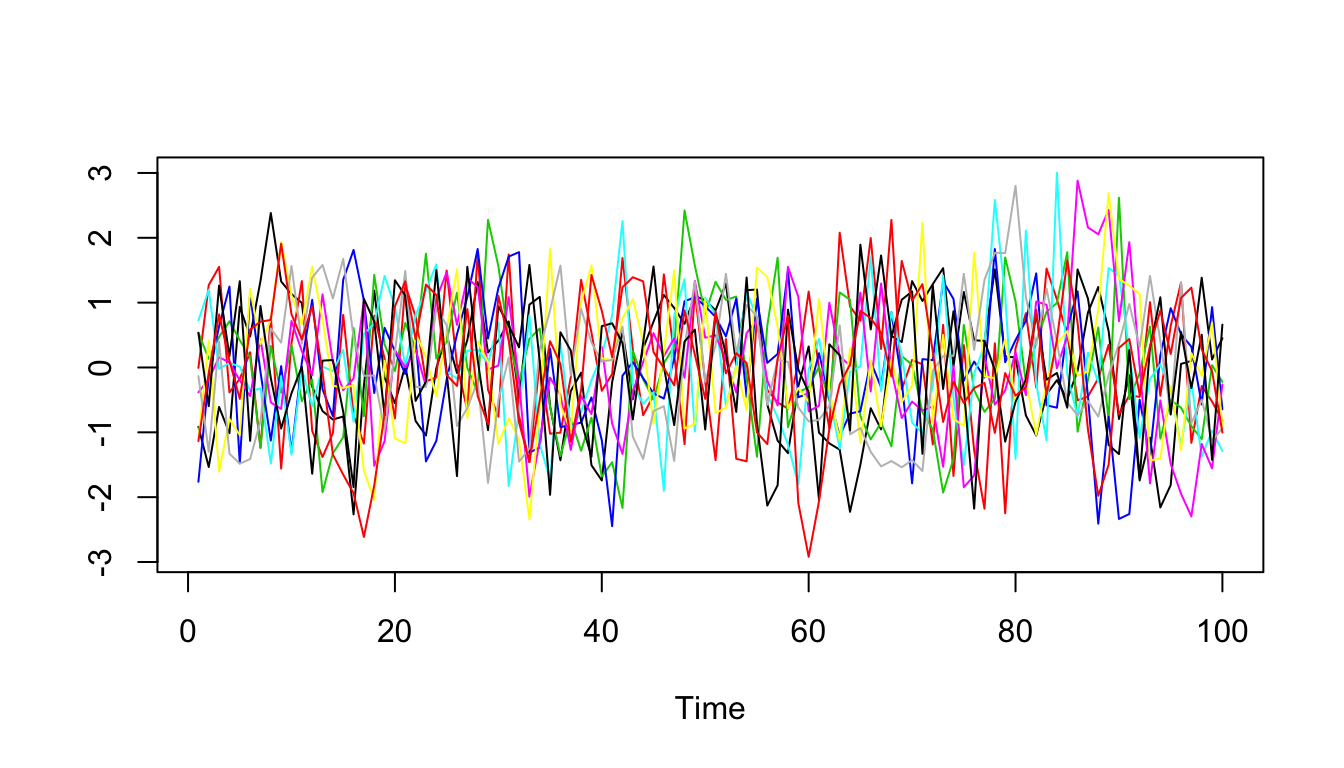
IC1
par(mfrow=c(1,2))
ts.plot(scale(A.est[,1]),col=1:10,ylab='')
map1=array(scale(re$S[,1]),dim=dims[1:3])
image(map1[,,10],breaks=c(-100:100)/100*21,col=bluered(200),xaxt='n',yaxt='n')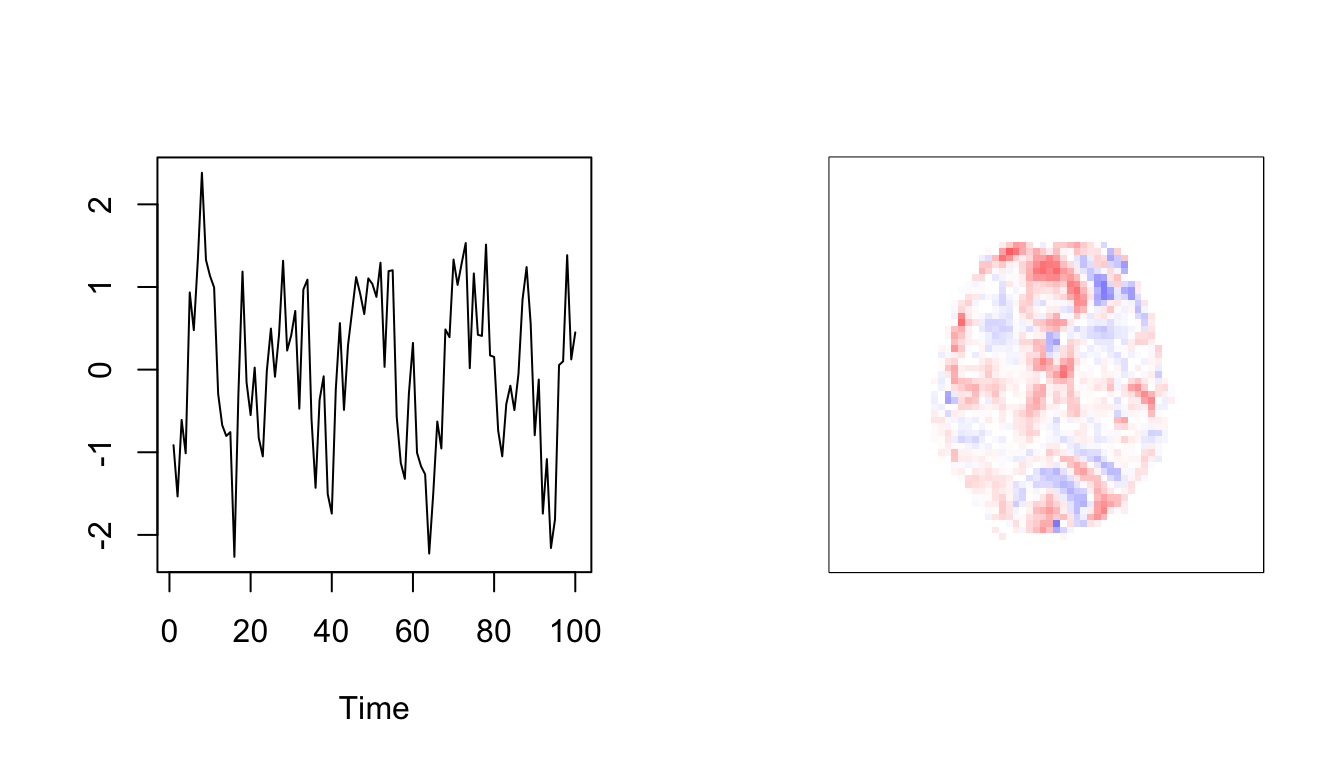
IC5,10
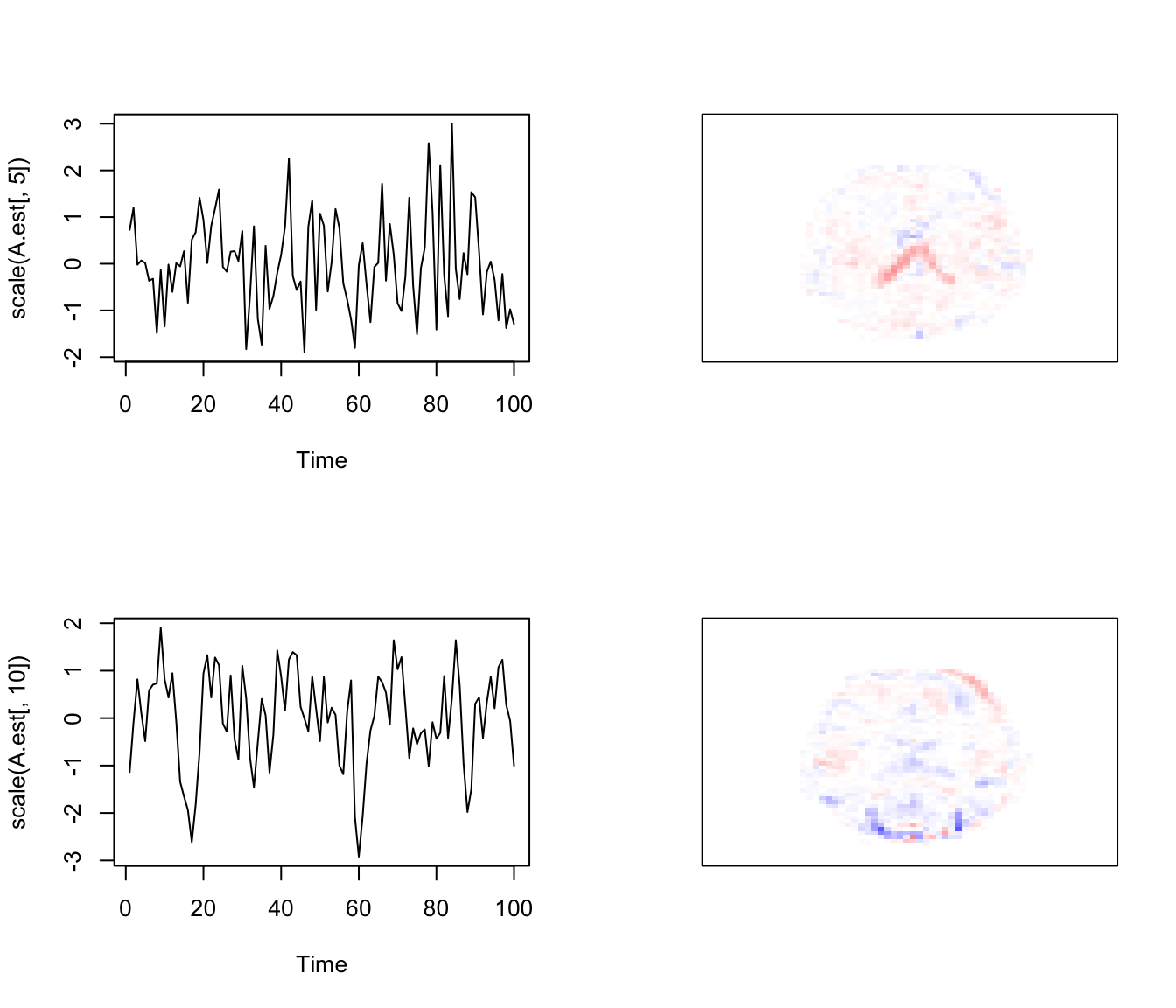
Some note.
fastICA has built in prewhitening!
Unfortunately, it won’t give you exactly same answer because the algorithm is stochastic! (due to optimization)
A lot of ICA algorithms are designed to have random initialization (like K-means clustering). They often gives similar answers, but not always. Have to be cautious.
library(fastICA)
set.seed(1235)
system.time(re2<-fastICA(t(X.c),n.comp=10))
image(cor(cbind(re$S, re2$S))[1:10,1:10+10],
col=bluered(200),breaks=c(-100:100)/100)Conclusion
- We will discuss group ICA and advanced ICAs (longitudinal ICA) next week!
